intro to bioinformatics: 16S rRNA microbiome profiling
View the workshop code on Github
Objective
Modify and update an existing workshop on introductory bioinformatics & microbial ecology to include more current tools and skills in the field.
Background
The workshop was originally designed as a two-hour class presented to undergraduate microbiology students. It included some background information on microbial ecology, and utilized data from Filippo et al. (2010) in order to compare the taxonomic differences between microbiota in the guts of children on a European diet versus children from Africa.
The original workshop utilized the online classifier tool provided by the Ribosomal Database Project. After data was generated online, students used Microsoft Excel to clean, calculate and visualize their data.
Methods
The original background information and data was kept, but the data processing steps were reworked to focus on command line and programming-based tools. Students used command line to clone the class files from Github, preview fasta files and count the number of fasta entries in a file. They then used the RDP classifier via command line to generate taxonomic and comparative tables that were then visualized using RStudio.
Results
Below are the figures generated by students:
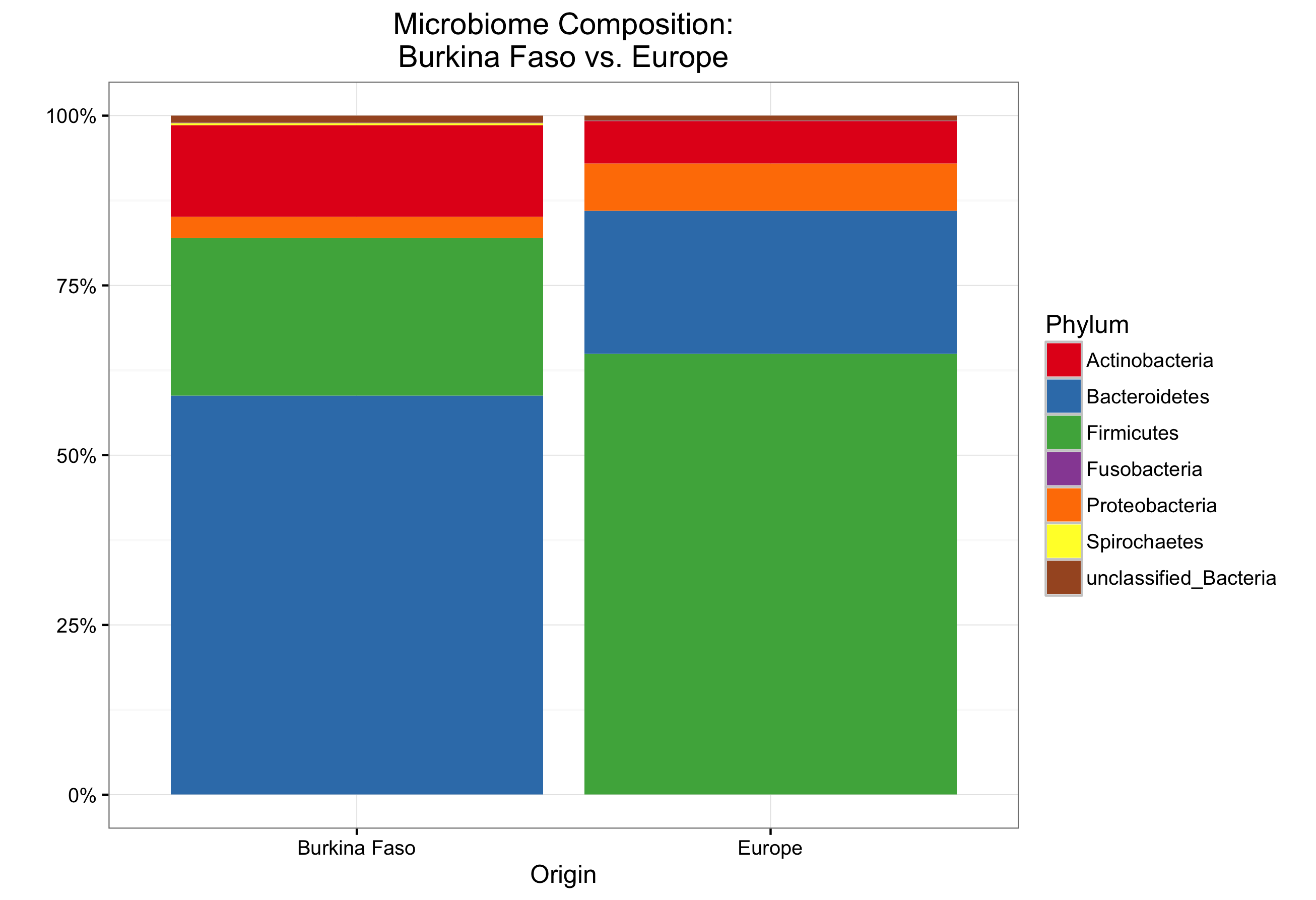
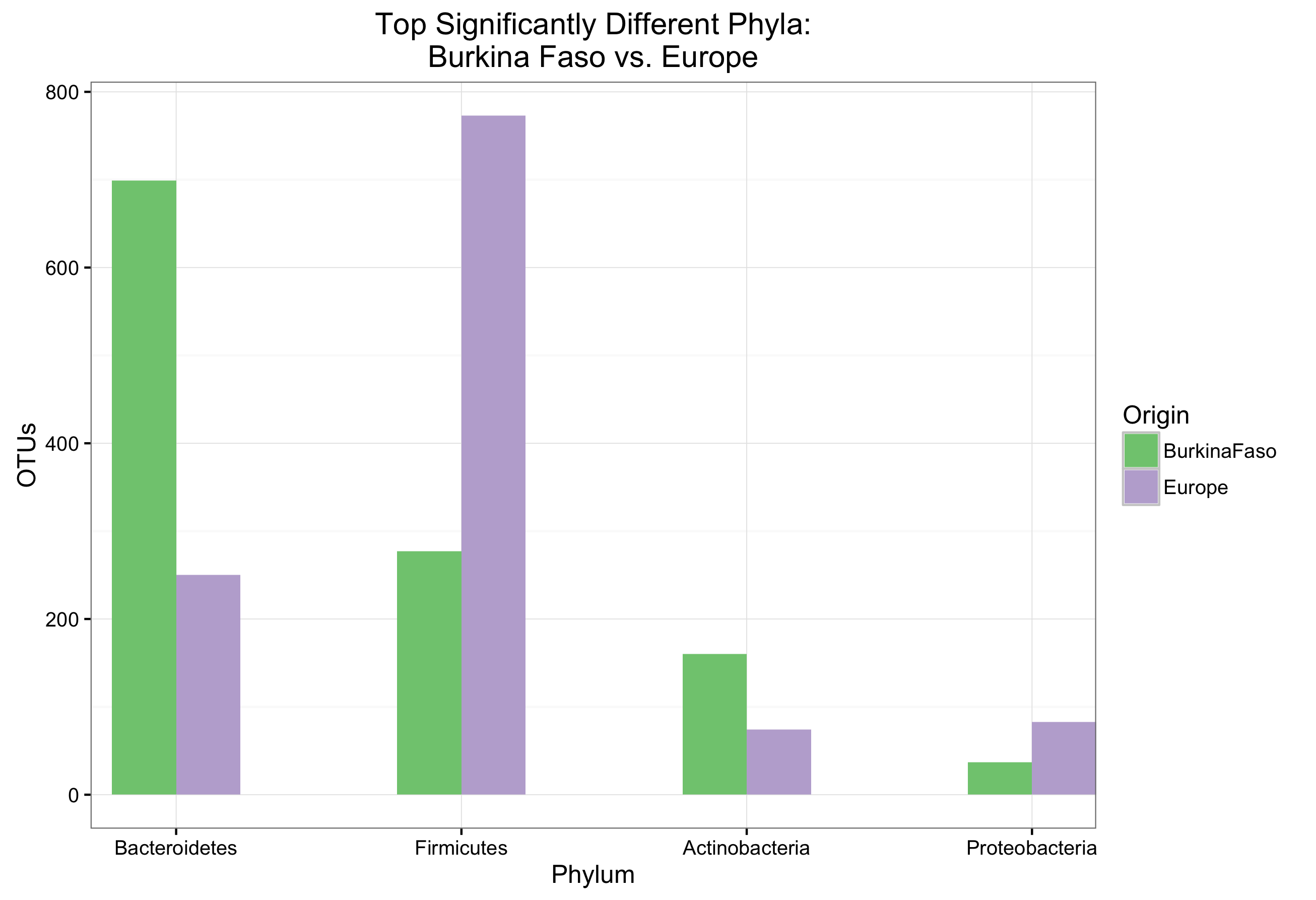
The course was presented to two lab groups, and all students were able to complete the assignment in time. Overall it was very well received. I believe one reason why the course worked so well was that the laptops provided to the students for the exercise were all a unix-based operating system, with the necessary programs and R libraries pre-installed.
Future
I provided the course material on Github so that in the future other graduate students can modify and present this same workshop. In the future I think some tweaks to the format would be ideal, including the use of a jupyter notebook, as well as a webpage presenting the material.
